Visualizing the Mechanics of Hormone-Driven Gene Activation: Insights into Breast Cancer
Scientists at Nano Life Science Institute (WPI-NanoLSI), Kanazawa University have captured real-time footage showing how a key hormone receptor activates genes, offering a clearer view into one of the most fundamental Kaprocesses in biology.
Using high-speed atomic force microscopy (HS-AFM), Richard Wong and colleagues at Kanazawa University directly visualized how the estrogen receptor alpha (ERα) binds to DNA and switches on genes in response to the hormone estrogen. Their findings, published in ACS Nano, reveal new molecular details of hormone signaling, with important implications for diseases like breast cancer.
Key Points
- Researchers used high-speed atomic force microscopy (HS-AFM) to directly film how estrogen receptor alpha (ERα) binds to DNA in living-like conditions.
- Estrogen binding improves ERα’s ability to dimerize and accurately target specific DNA sites called estrogen response elements (EREs).
- Findings support a new ‘Ligand-Induced Dimerization (LID)’ model and offer insights into hormone-driven diseases like breast cancer.
Estrogen receptors play a critical role in controlling gene activity in many tissues. When estrogen binds to ERα, the protein changes shape, forms a dimer (a molecular pair), and attaches to specific regions of DNA called estrogen response elements (EREs). Although the importance of this process has been known for decades, it had never before been observed unfolding at the single-molecule level in real time.
To capture this, the researchers used HS-AFM to scan individual ERα molecules interacting with DNA. They compared the behavior of ERα with and without estrogen present. Their experiments showed that ERα could bind to DNA without estrogen but did so less precisely and less stably. When estrogen was present, ERα molecules dimerized more efficiently and exhibited targeted, stable binding to ERE sequences.
“Our study shows that estrogen acts like a molecular matchmaker,” says Richard Wong. “It not only triggers ERα to find the right DNA sequence but also stabilizes its grip, ensuring accurate gene activation.”
Based on these observations, the team proposed a new ‘Ligand-Induced Dimerization’ (LID) model explaining how hormones fine-tune the dynamic behavior of receptors at the DNA level.
This work provides direct visual evidence of how molecular signals from hormones lead to precise gene control — a fundamental advance that could guide new strategies for treating hormone-driven diseases.
Glossary
- Estrogen receptor alpha (ERα): A protein inside cells that binds estrogen and controls the activity of specific genes.
- High-Speed Atomic Force Microscopy (HS-AFM): An imaging technology that captures the movement of individual molecules in real time.
- Ligand: A molecule, such as estrogen, that binds to a protein and changes its shape or function.
- Dimerization: The process by which two protein molecules join together to form a pair (dimer), often necessary for function.
- Estrogen Response Element (ERE): A specific sequence of DNA that the estrogen receptor recognizes and binds to, controlling gene expression.
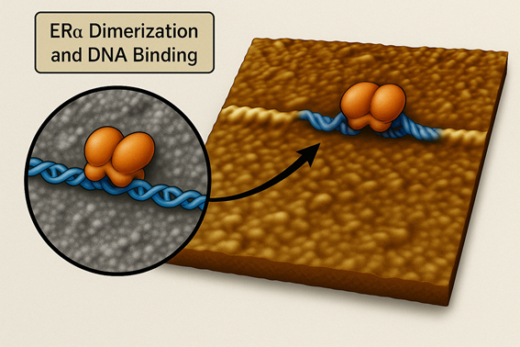
Figure: How Estrogen Receptor Binds DNA.
This illustration shows the estrogen receptor alpha (ERα, in orange) attaching to DNA (in blue) as a pair, or dimer. The image is based on real-time, high-speed atomic force microscopy (HS-AFM) data, showing how ERα recognizes and binds specific DNA sequences to activate genes. The close-up highlights the dimer sitting on the DNA strand — a key step in hormone-driven gene regulation.
Article
- Title
- Zooming into Gene Activation: Estrogen Receptor α Dimerization and DNA Binding Visualized by High-Speed Atomic Force Microscopy
- Author
- Goro Nishide, Tomoka Ishibashi, Keesiang Lim, Yujia Qiu, Masaharu Hazawa, Ayami Matsushima#, Richard W. Wong#
- Journal
- ACS Nano
- Publication date
- Apr 18, 2025
- DOI
- 10.1021/acsnano.4c14943
- URL
- https://doi.org/10.1021/acsnano.4c14943

