Discovering the molecular recognition of technological solids by mutant self-assembling peptides
In a study recently published in the journal SMALL, a weekly peer-reviewed scientific journal covering nanotechnology, published by Wiley-WCH, Germany, researchers from Nano Life Science Institute (WPI-NanoLSI), Kanazawa University, Japan, collaborating with Professor Sarikaya, Seattle, USA, used frequency modulated atomic force microscopy to reveal the molecular architecture of genetically designed and point mutated peptides and their self-organizations each forming single-molecule thick, distinct biological crystals on atomically flat graphite and MoS2 surfaces, offering a potential platform for hybrid technologies such as bioelectronics, biosensors, and protein arrays.
Proteins are the key molecular building blocks in biology, carrying out enzymatic functions, transporting ions and electrons, and constituting the major infrastructure of cellular architectures, such as ion pumps. The functions of proteins depend on their amino acid sequences, which, in turn, dictate their three-dimensional molecular structures. The amino acid sequences of proteins are coded by the genes (DNA code) of a given specialized cell. Any defect in the amino acid sequence, such as a point mutation, a change in the location of an amino acid, a missing amino acid, or the transposition of two or more amino acids, is therefore expected to affect the molecular structure of the protein, which is reflected in their function, often leading to genetic diseases.
Similar to proteins, peptides are also composed of amino acid units, and each peptide has a unique sequence; however, they are much shorter, ranging from 10 to 30 units, and similarly coded by DNA. They are easier to manipulate, design, and use compared to proteins, which are much larger (hundreds or thousands of amino acids long), difficult to synthesize, and impractical to use. Therefore, it is imperative that if practical technology of the future is to be developed based on lessons from biology at the molecular scale, the fundamental science has to be established using peptides, which may then be implemented in real-world applications, such as biosensors, bioelectronics, and logic devices, as well as drugs, all based on lessons from biology. Realizing this premise, Professor Sarikaya initiated a novel convergence science approach two decades ago, involving genetic engineering, molecular biology, informatics, machine learning, and materials science and engineering, in designing solid binding peptides, and demonstrated their use in practical hybrid technologies.
Here, in this study, one of these peptides, specifically designed as a graphite-binding peptide, is mutated into two different peptides, M6 and M8, each nominally the same molecular weight and size but containing point mutations of the charged amino acids, one negative and the other positive, respectively. As predicted, they would function differently, which they did. However, the question that was impossible to predict a priori was how exactly they would act differently from each other and the wild type (the original) peptide, and what would be the details of their molecular structure and their kinetic behavior on the surface of the substrate graphite. The answers to these questions, the lack of which hitherto limited the understanding of the relationship between mutants and their predictable behavior, required very high resolution and direct imaging of the molecules at their native states, in water over a period of seconds to hours. This is where Professor Takeshi Fukuma’s Laboratory comes into the picture at WPI Nano Life Science Institute, Kanazawa University, where his team has developed Frequency-Modulated Atomic Force Microscopy facilities that are capable of interrogating molecular-scale objects at near-angstrom image resolution and for long periods of time within aqueous environments, an unusual combination of capabilities for an experimental system, especially for biological research.
With the highly proficient experimental abilities of the researchers (with inexhaustible patience in long-duration tests), led by Dr. Ayhan Yurtsever, Dr. Kaito Hirata, Dr. Linhao Sun, and Dr. Takeshi Fukuma, the team discovered the behavior of the mutants in unprecedented detail, providing results with significance that crosses several traditionally diverse disciplines, from materials science to physics, to chemistry, and biology. Firstly, despite the mutations and individually having a different overall charge, each peptide not only bound to the graphite surface but also assembled, forming single-molecule-thick peptide crystals. However, while the first mutant, the negatively charged one, formed a crystalline oblique lattice with two molecules (wrapped around each other) in each lattice, the second mutant, the positively charged one, formed a different oblique lattice containing a single peptide. What is truly valuable here is that the behavior of the peptides is simply and directly related to their molecular architecture, modeled using molecular dynamics. The researchers, therefore, were able to mathematically describe the relationship between each of the peptides (each forming a different oblique molecular lattice) and the hexagonal lattice of the graphite substrate, which is exclusively described as chiral recognition. In effect, this means that each peptide is demonstrated to form a hybrid interface with the substrate graphite, best described as being crystallographically coherent with no noticeable gap in between, not unlike ligand-receptor or protein/DNA interactions in biology.
Neither the molecular recognition of solid-state objects by a ‘living’ molecule has been so clearly observed before, nor have its details been described as chiral, both adding to the significance of the results presented in this publication. On the one hand, from the biology perspective, there is now ample data and details on how mutations lead to molecular conformational changes and the resultant function of the peptides, which can be expanded into the behavior of proteins, with significance in genetic mutations and effective drug design. And on the other hand, coherent hybridization suggests that animate and inanimate nanoscale objects are stably present at the same soft interface, forming an exclusive molecular bridge that could be best described by seamlessly integrating biology with solid-state devices. If so, based on this study, scientists and technologists are expected to enormously widen the repertoire of the molecular basis of future bio/nano devices by using the simple molecular biology tool of mutation.
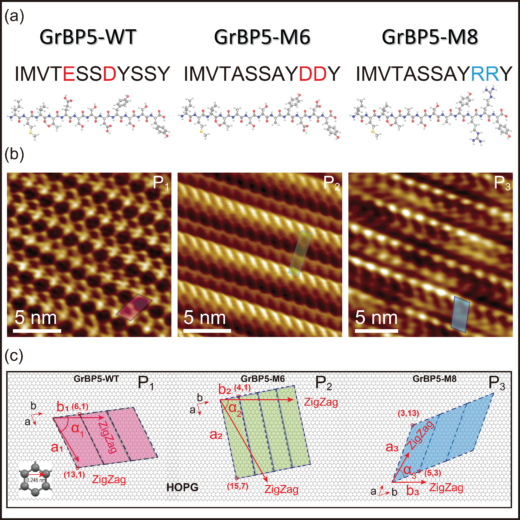
Figure 1: Molecular characteristics and self-organization of the peptides:
(a) Basic chemical data of GrBP5-WT and its two mutations, GrBP5-M6 and GrBP5-M8.
(b) Ordered organization of the peptides at high resolution, revealing the details of the lattice structures.
(c) Crystallographic orientation relationships between the single molecule-thick peptide crystals and surface lattice of cleaved HOPG that exposes (0001) lattice of carbon atoms on atomically flat graphite are determined by lattice matching which shows an exclusive chiral relationship for each peptide.
© 2024 Yurtsever, et al., Small published by Wiley-VCH GmbH
Article
- Title
- Dynamics of Molecular Self-Assembly of Short Peptides at Liquid–Solid Interfaces – Effect of Charged Amino Acid Point Mutations
- Author
- Ayhan Yurtsever, Kaito Hirata, Ryohei Kojima, Keisuke Miyazawa, Kazuki Miyata, Sanhanut Kesornsit, Hadi Zareie, Linhao Sun, Katsuhiro Maeda, Mehmet Sarikaya, Takeshi Fukuma
- Journal
- Small
- Publication date
- Feb 22, 2024
- DOI
- 10.1002/smll.202400653
- URL
- https://doi.org/10.1002/smll.202400653

