Ion channel block unraveled
Researchers at Kanazawa University report in Nature Communications how calcium ions can block sodium ion channels located in cell membranes. Structural analysis and computer simulations made it possible to identify where and why calcium ions get stuck.
Ion channels are structures within cell membranes that enable specific ions to travel to and from the cell. Such transfer is essential for a variety of physiological processes like muscle cell contraction and nerve excitation. In so-called tetrameric cation channels, the ion selectivity results from the unique structural and chemical environment of the part referred to as the selectivity filter, which is located between two intertwined helical structures. Tetrameric ion channels are prone to ‘divalent cation block’, the blocking of the channel by ions like calcium (Ca2+). Such blocking regulates the ionic current, which is involved in various neural activities such as memory formation. How exactly divalent cation block happens is still unclear at the moment — particularly, a direct observation of the cation actually blocking the ion pathway has not been reported yet. Now, Takashi Sumikama from Kanazawa University in collaboration with Katsumasa Irie from Wakayama Medical University and colleagues has discovered the mechanism behind divalent cation block in NavAb, a well-known tetrameric sodium (Na) channel. Through structural analysis and computer simulations, the researchers were able to reveal the relevant structural features and molecular processes at play.
NavAb is a sodium channel cloned from a bacterium (Arcobacter butzleri) and has a well-known structure. Sumikama and Irie’s colleagues performed experiments with NavAb and three mutants. The structures of the mutants were determined for environments with and without calcium. The scientists focused on the differences in electron densities for the different structures, as these provide insights into the locations of the calcium ions. They found that for the mutants displaying calcium blocking, one or two calcium ions are located at the bottom of the selectivity filter. They also discovered that magnesium (Mg2+) and strontium (Sr2+) ions, two other divalent cations, blocked the calcium-blocking mutant sodium channels.
The researchers then performed computer simulations to obtain a detailed understanding of the interaction between the calcium ions and the mutated NavAb channels. The simulations reproduce the dynamics of ions passing — or not passing — the channel. In the absence of calcium ions, sodium ions were observed to penetrate the channel. In the presence of calcium ions, penetration significantly decreased in the calcium-blocking mutants. The simulations also confirmed that the blocking calcium ions are ‘stuck’ at the bottom of the selectivity filter, and revealed that this ‘sticking’ is related to the increased hydrophilicity (affinity to water) of relevant structural parts of the mutated channels.
The results of Sumikama and Irie’s colleagues provide an important step forward towards a full understanding of the mechanism of divalent cation block in NavAb, an important and representative sodium ion channel. Quoting the scientists: “… our results and methods of structural analysis and molecular dynamics simulations are … expected to play an active and meaningful role in the advanced analysis of divalent cation-blocking mechanism.”
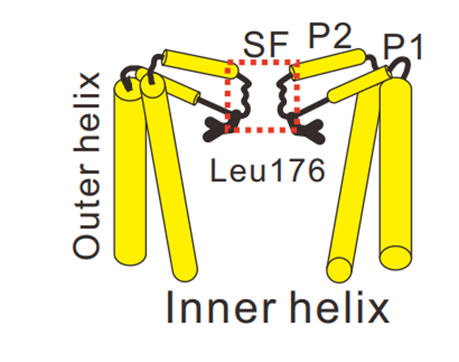
Figure 1. Schematical illustration of the pore domain of a tetrameric cation channel, with the selectivity filter (SF) located between two transmembrane helices (Outer and Inner helix). ©2023 Irie, et al CC BY 4.0

Figure 2. Representative coordinates of waters and calcium ions around the P1-helix C-termini and the proposed molecular model of divalent cation block.
Background
Ion channels are proteins embedded in cellular membranes enabling particular ions to pass through the membrane. They are present in the membranes of all types of cells. The regulation of ion transfer across cellular membranes — in particular, the flow of calcium, potassium and sodium ions —is essential for many physiological processes, including insulin release, heart muscle contraction and nerve signaling. Ion channels enable passive ion flow towards equilibrium, which is driven by ion concentration differences across the membrane and the membrane potential (the difference electric potential between the cell interior and exterior).
Tetrameric cation channels are the largest and best characterized group of ion channels. For this type of ion channel, the ion pathway lies in a pore domain consisting of two transmembrane helices surrounding the so-called selectivity filter — a unique structural and chemical environment that lets only one species of ion pass. In tetrameric cation channels, a phenomenon known as divalent cation block is known to occur: the blocking of ion transport due to the presence of divalent cations such as calcium (Ca2+) or magnesium (Mg2+). The mechanism underlying cation block is not well understood. But now, by combining structural studies and computer simulations of a bacterium-based tetrameric sodium channel and mutated variants, Takashi Sumikama from Kanazawa University, Katsumasa Irie from Wakayama Medical University and colleagues provide valuable mechanistic insights into the origins of divalent cation block.
Article
- Title
- The structural basis of divalent cation block in a tetrameric prokaryotic sodium channel
- Author
- Katsumasa Irie, Yoshinori Oda, Takashi Sumikama, Atsunori Oshima, and Yoshinori Fujiyoshi.
- Journal
- Nature Communications
- Publication date
- Jul 15, 2023
- DOI
- 10.1038/s41467-023-39987-0
- URL
- https://www.nature.com/articles/s41467-023-39987-0

