Direct visualization of dynamic structures of protein disaggregation molecular machines
Abstract:
ClpB, an ATP-fueled protein molecular machine, disentangles and reactivates aggregated proteins. By using high-speed atomic force microscopy, conformational dynamics of ClpB was visualized for the first time. ClpB forms open and closed rings. The closed ring was observed to take three forms; round, spiral, and twisted-half-spiral forms. These structures were observed to transform from one to another during the ATPase-cycle, indicating that ClpB performs protein disaggregation through these structural changes. In the future, this result will contribute to establishing a remedy for various protein aggregation-related diseases.
Background
When proteins are exposed to stresses such as a heat shock, they lose their native structure and form toxic insoluble aggregates. Bacterial molecular chaperone ClpB and its yeast homologue Hsp104 have an ability to disentangle and reactivate aggregated proteins. The three-dimensional structure of ClpB/Hsp104 has been determined by X-ray crystallography and cryo-electron microscopic single-particle analysis. ClpB consists of two ATPase cores, AAA1 and AAA2, and two additional domains, the N-domain and the M-domain. ClpB assembles to form a hexameric ring. Using the chemical energy of ATP hydrolysis, the ClpB ring threads an aggregated protein through its central pore to disentangle it. The disaggregation activity is regulated by the rod-shaped M-domains surrounding the ring periphery. Despite these known aspects, it has not been unraveled how ATP binding and hydrolysis change the ClpB structure and how structural changes are related to the disaggregation activity. To answer these central questions, we needed to observe individual ClpB rings in dynamic action.
Results
By using high-speed atomic force microscopy (HS-AFM), this research group succeeded for the first time in observing structural changes occurring in individual ClpB rings with 100-ms temporal resolution. In the presence of ATP, ClpB forms both closed and open rings (Fig. 1). The number of subunits contained in a ring and the ring polymorphism were further confirmed by native mass spectrometry and negative-staining electron microscopy analysis, respectively. During HS-AFM observation, these two conformations converted from one to another. The population of the closed ring increased with increasing ATP concentration. Moreover, closed rings displayed distinct three forms: the “round form” whose height is almost uniform, the “spiral form” in which the height changes continuously like a spiral staircase, and the “twisted-half-spiral form” in which two half-spiral units face each other (Fig. 1). The twisted-half-spiral form suggested that the hexameric ring would consist of two trimers, which was supported by sedimentation velocity analytical ultracentrifugation. These three distinct forms also converted from one to another. The conversion frequency became higher with increasing ATP concentration. These observations revealed that the ATP binding induces the closed ring formation, while ATP hydrolysis causes massive structural changes between the round, spiral, and twisted-half-spiral forms.
From the HS-AFM observations of ClpB mutants whose ATP binding, ATP hydrolysis on either side of the AAA1 and AAA2 domains or ATP hydrolysis on both domains was inhibited, individual roles of these two domains in the structural dynamics were clarified. ATP binding to the AAA1 domain induces oligomerization of ClpB, and the hexameric state is stabilized by ATP binding to the AAA2 domain. The structural changes between the round, spiral, and twisted-half-spiral forms are caused by ATP hydrolysis on the AAA2 domain (Fig. 2). Moreover, the structural changes of the closed ring were drastically decreased in an M-domain mutant that lost disaggregation activity but retained ATPase activity. Thus, the structural changes play an important role in the disaggregation reaction.
Future prospects
Protein aggregations are closely related to various diseases, including Alzheimer’s and Huntington’s diseases. The formation of protein aggregations is also problematic in the use of proteins in medical and industrial fields. The results of this research have a potential to contribute to treating these diseases and maintaining useful proteins. Furthermore, ClpB belongs to the AAA+ protein family consisting of various important proteins contributing to DNA replication, membrane fusion, protein degradation, circadian clock maintenance and others. The members of this family share AAA+ domains as ATPase cores. Therefore, the results of our present study are expected to lead to the elucidation of a common mechanism that these AAA+ family proteins share.
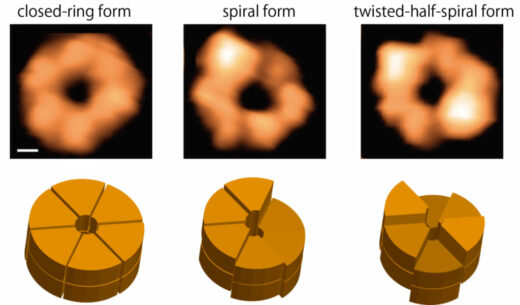
Figure1. Ring forms of ClpB
This figure shows snapshots (upper panels) of the closed ring of ClpB captured by HS-AFM and their schematics (lower panels). The closed ring structure is classified into the round, spiral and twisted-half-spiral forms. Scale bar in the upper left panel, 2 nm.
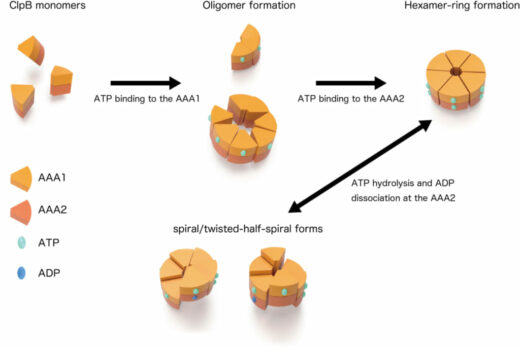
Figure2. Dynamic changes of ring structure of ClpB
Schematic of structural conversions in the closed ring of ClpB. The ring structure is converted to distinct forms in an ATPase-cycle dependent manner.
Article
- Title
- Dynamic Structural States of ClpB Involved in Its Disaggregation Function
- Author
- Takayuki Uchihashi†, Yo-hei Watanabe†*, Yosuke Nakazaki, Takashi Yamasaki, Hiroki Watanabe, Takahiro Maruno, Kentaro Ishii, Susumu Uchiyama, Chihong Song, Kazuyoshi Murata, Ryota Iino*, Toshio Ando* (†co-first authors, *corresponding authors)
- Journal
- Nature Communications
- Publication date
- Jun 1, 2018
- DOI
- 10.1038/s41467-018-04587-w
- URL
- https://www.nature.com/articles/s41467-018-04587-w
Funder
・Grant-in-Aids for Scientific Research to Takayuki Uchihashi (JP15H03540, JP16H00830, and JP16H00758)
・Grant-in-Aid for Scientific Research to Yo-hei Watanabe (JP26440085)
・Grant-in-Aids for Scientific Research to Susumu Uchiyama: JP16H00770 and JP17H03975)
・Grant-in-Aids for Scientific Research to Ryota Iino (JP15H04366, JP16H00789, JP16H00858, and JP17K19213)
・Grants-in-Aids for Scientific Research to Toshio Ando (JP24227005 and JP26119003)
・Support to Susumu Uchiyama from BIONEXT project in the Okazaki Institute for Integrative Bioscience, National Institutes of Natural Sciences
・Grant from JST/CREST to Toshio Ando (JPMJCR13M1).

