Fate of human cells: visualizing gene expression and cell function with innovative live cell imaging technology
Yusuke Miyanari, Associate Professor, Nano Life Science Institute (WPI-NanoLSI), Kanazawa University
The Human Genome Project was successful in mapping the entire genome sequence, and recent research has revealed the chemical makeup of (epigenetics) of chromatin–bead-like strings composed of DNA and proteins constituting chromosomes within the nucleus of human cells. However, the regulatory mechanisms governing gene expression, and importantly, why cells that share the same DNA sequences evolve to perform differing functions still not fully understood. For example, skin cells and those constituting heart muscles perform different tasks even though they originate from the same stem cells.
In order to contribute to this area of research, Yusuke Miyanari, who joined the Life Science group of the WPI NanoLSI at Kanazawa University in April 2020, is studying molecular mechanisms governing cell fate decisions. He is focusing on unraveling the higher order structures of chromatin to gain insights into control mechanisms of gene expression. This research is expected to lead to insights into causes of diseases such as cancer, which are related to irregular fluctuations in gene expression that govern the growth of all cells constituting the human body.
From pharmacy in Kyushu to visualizing mechanisms for virus replication in Kyoto
“I have a bachelor’s degree in pharmacy from Kyushu University,” says Miyanari. “Frankly, there was no special reason for studying pharmacy; I just did it. But after my undergraduate studies I was faced with the option of three main career paths. Work for a pharmaceutical company, become a pharmacist and set up my own pharmacy or become a research scientist. I chose scientific research and started my doctorate at Kyoto University on hepatitis C virus replication. My doctoral research forms the foundation for all of my research activities to date.”
One of the papers published by Miyanari for his doctorate reports on the important role of lipid droplets—highly specialized structure within living cells—in the assembly of hepatitis C virus particles [1]. A combination immunofluorescence imaging and electron microscopy revealed that “lipid droplets were involved in the production of infectious virus particles”. This study showed how viruses can take control of cellular functions.
Research on nuclear dynamics in Japan and France
After completing his doctorate Miyanari moved to the National Institute of Genetics (NIG) in Mishima, Shizuoka Prefecture, Japan. One of the highlights of his research at NIG was a paper on methylation-specific fluorescence in situ hybridization (MeFISH) for microscopic imaging of “DNA methylation status at specific repeat sequences in individual cells” [2]. “My research at NIG led to important advances in detecting specific hypomethylated satellite repeats in human cells,” explains Miyanari. “Since the publication of this paper in 2013, our technique of MeFISH has been extensively used in epigenetics as well as medical diagnostics.”
In 2009 Miyanari moved to the Maria-Elena Torres-Padilla Lab at the Institute of Genetics and Molecular and Cellular Biology (IGBMC) in Strasburg, France. “My five years at IGBMC enabled me to contribute to research on pluripotency [3] and the development of transcription activator–like effector (TALE) mediated genome visualization (TGV) to label specific sequences of the genome for tracking chromatin dynamics in live mouse cells [4],” explains Miyanari. “This was a major advance in visualizing nuclear dynamics. I also enjoyed seeing at first-hand differences in approaches to conducting research between Japan and France. For example, in my lab people were careful to take the minimum of data and submit to a high impact journal, which contrasts with Japan where people take a lot of data to cover potential questions from referees. I also found the atmosphere at IGBMC to be free of hierarchy, with excellent interaction between scientists, administrators, and technical staff.”
More recently, Miyanari and colleagues developed APEX-mediated chromatin labeling and purification (ALaP) to visualize and track the exact location of promyelocytic leukemia structures that are closely associated with chromatin bodies on the chromatin [5,6].
Visualizing hundreds of proteins and harnessing the power of NanoLSI-technology
One of the challenges in this field of research is the huge number of proteins involved in the process of expressing a single gene. For example, it takes more than 100 proteins to express a single gene, but it is only possible to image about four proteins using even the most advanced imaging technology. “We have realized that only being able to distinguish and label a handful of proteins is not sufficient to fully understand the control mechanisms of gene expression,” says Miyanari. “My approach is to produce ‘unusual’ proteins, for example from bacteria, and collaborate with people who have expertise in imaging, to develop tools to observe my unique proteins that are used as labels. For example, in the TGV experiments, we used a conventional fluorescence microscope, but the important point is that the observations were made with my unique protein labels.”
To-date, Miyanari has based his research on fluorescence microscopy and genetic sequencing. At the NanoLSI he wants to collaborate with the scientists to design experiments to exploit the power of AFM-based technology to shed light on the mysteries of gene expression and cell fate determination.
“Deeper knowledge about the mechanisms responsible for controlling gene expression will enable us determine the causes of human ailments triggered by abnormal gene expression such as tumorigenesis, virus infections, and developmental defects. Such powerful insights will offer new opportunities to develop drugs to mitigate life threatening diseases.”
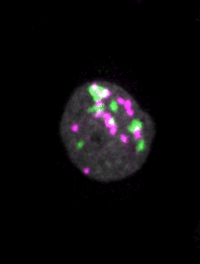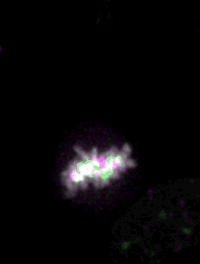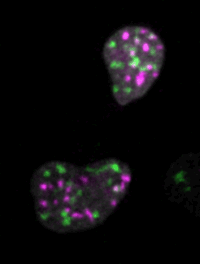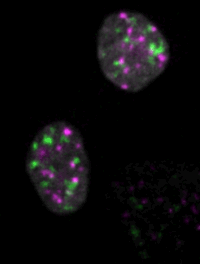
Live imaging of mouse ES cells tells us how nuclear dynamics contributes to the cell-fate decision.
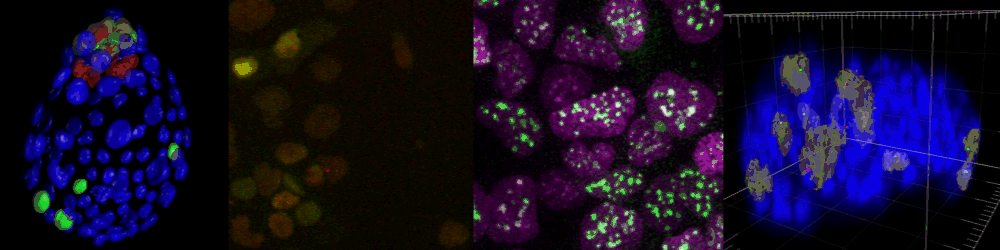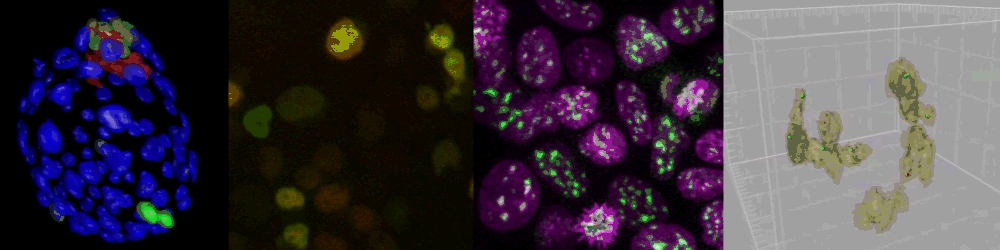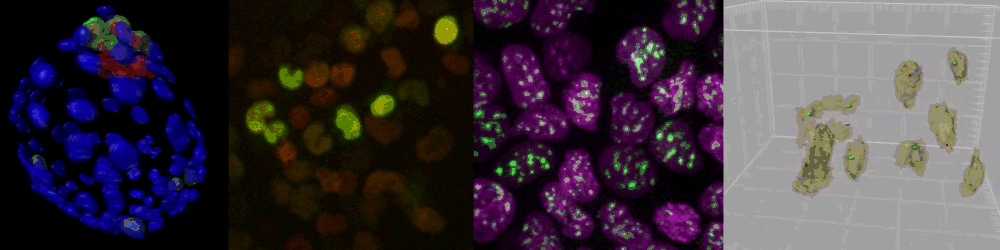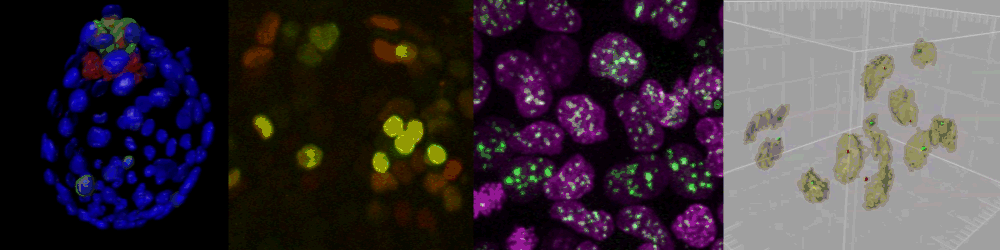
Several imaging techniques boost their studies.
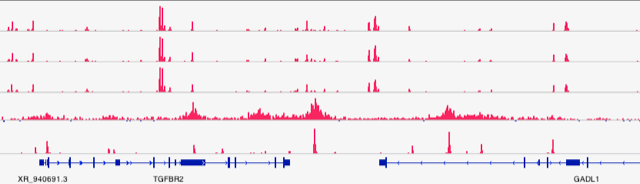
Deep sequencing shed light on chromatin regulation at molecular level.
References
[1] Yusuke Miyanari et al, The Lipid droplet is an organelle important for Hepatitis C virus production, Nature Cell Biology, 9, 1089–1097 (2007).
DOI: 10.1038/ncb1631
[2] Yufeng Li et al, Sequence-specific microscopic visualization of DNA methylation status at satellite repeats in individual cell nuclei and chromosomes, Nucleic Acids Res, 2013 Oct;41(19):e186.
DOI: 10.1093/nar/gkt766
[3] Yusuke Miyanari & Maria-Elena Torres-Padilla, Control of ground-state pluripotency by allelic regulation of Nanog, Nature 483, 470–473, (2012).
DOI:10.1038/nature10807
[4] Yusuke Miyanari, Live visualization of chromatin dynamics with fluorescent TALEs, Nature Structural & Molecular Biology, 20, 1321–1324, (2013).
DOI: 10.1038/nsmb.2680
[5] Misuzu Kurihara et al, Genomic Profiling by ALaP-Seq Reveals Transcriptional Regulation by PML Bodies through DNMT3A Exclusion, Molecular Cell, 78, 493-505.E8, 2020.
DOI: 10.1016/j.molcel.2020.04.004
[6] WPI-NanoLSI Press Release
Dock and Harbor: A Novel Mechanism for Controlling Genes
https://www.kanazawa-u.ac.jp/latest-research/79753
[7] Ishii S. et al, Genome-wide ATAC-see screening identifies TFDP1 as a modulator of global chromatin accessibility
Nature Genetics. 2024 Mar;56(3):473-482.
DOI: 10.1038/s41588-024-01658-1

