AFMnanoSALQ: A Novel HS-AFM Image Analysis Framework
Summary
Researchers at Department of Computer Vision and Cognitive Cybernetics, Faculty of Information Technology, University of Science, Vietnam National University Ho Chi Minh and Nano Life Science Institute (WPI-NanoLSI), Kanazawa University have successfully developed a novel HS-AFM data analysis framework: AFMnanoSALQ: An Accurate Detection Framework for Semi-Automatic Labeling and Quantitative Analysis of α-Hemolysin Nanopores Using Intensity-Height Cues in HS-AFM Data.
AFMnanoSALQ serves as a practical tool for preliminary data inspection, capable of independently performing instance boundary detection and quantitative morphological measurement, while also facilitating semi-automatic labeling to accelerate the creation of training datasets. The framework provides a robust foundation for future deep-learning studies by supporting both efficient dataset generation and cross-validation between feature-driven and data-driven approaches.
In this study, α-Hemolysin (αHL), a pore forming toxin from Staphylococcus aureus (PDB: 7AHL), serves as a representative and challenging case due to its distinct mushroom-shaped and dynamic prepore and pore-like conformations, which are highly compatible with feature-based analysis. Although AFMnanoSALQ is optimized for analysis of αHL images, the framework is adaptable and can be extended to other biomolecular systems with appropriate feature-extraction customization.
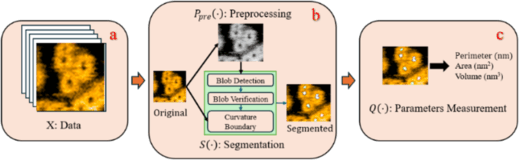
Scheme 1 Overview of AFMnanoSALQ for αHL HS-AFM analysis. (a) Dataset X comprises HS-AFM images capturing αHL dynamics over time. (b) P_pre (⋅) performs preprocessing (e.g., smoothing, grayscale converting, denoising) and S(⋅) detects instance boundary of “kernel” in conformations (e.g., oligomers, mushroom-shaped pre-pores, pores). (c) Q(⋅) computes morphological parameters (e.g., perimeter, area, volume, height). Together, these components form a comprehensive pipeline for quantitative analysis of αHL developmental stages in HS-AFM data (Copywirte © Springer Nature Singapore Pte Ltd 2026).
Main Contributions
- Feature-driven approach for HS-AFM data: AFMnanoSALQ integrates visual and geometric features to extract 3D morphology from AFM images.
- Independent instance analysis: AFMnanoSALQ can perform instance boundary detection and quantitative morphological measurement, enabling reliable preliminary data inspection.
- Semi-automatic labeling: AFMnanoSALQ accelerates the creation of training datasets for downstream deep-learning models, reducing the annotation burden.
- Application to α-hemolysin: AFMnanoSALQ effectively detects and measures αHL structures.
- Foundation for future deep-learning studies: AFMnanoSALQ supports cross-validation between feature-driven and data-driven approaches, providing high-quality datasets for downstream deep-learning applications.
Perspectives and future research
AFMnanoSALQ provides a robust framework for instance boundary detection and quantitative analysis of HS-AFM data. However, its performance depends on distinct structural features and well-preserved imaging quality. Our future research will focus on the following:
- Advanced deep-learning-based denoising to enhance robustness across diverse imaging conditions.
- Generalizing and validating feature-extraction modules on various biomolecules to broaden applicability.
- Leveraging AFMnanoSALQ-generated semi-automatic labels to train deep-learning models for downstream HS-AFM analysis tasks.
Article
- Title
- An Accurate Detection Framework for Semi-Automatic Labeling and Quantitative Analysis of α-Hemolysin Nanopores Using Intensity-Height Cues in HS-AFM Data
- Author
- Thuyen Tran Vinh Nguyen*, Ngoc Quoc Ly*, Ngan Thi Phuong Le, Hoang Duc Nguyen, Kien Xuan Ngo*. (*Corresponding authors)
- Journal
- Conference book: Multi-disciplinary Trends in Artificial Intelligence
- Publication date
- Nov 22, 2025
- DOI
- 10.1007/978-981-95-4963-4_7
- URL
- https://doi.org/10.1007/978-981-95-4963-4_7

