Sodium channel investigation
Researchers at Kanazawa University report in Nature Communications a high-speed atomic force microscopy(*1) study of the structural dynamics of sodium ion channels in cell membranes. The findings provide insights into the mechanism behind the generation of cell-membrane action potentials.
The transport of ions to and from a cell is controlled by pore-forming proteins embedded in the cell membrane. In particular, so-called voltage-gated sodium channels (VGSCs) govern the transfer of sodium (Na+) ions, and play an important role in the regulation of the membrane potential — the voltage difference between the cell’s exterior and interior. In electrically excitable cells such as neurons and muscle cells, VGSCs participate in the generation of action potentials; these are rapid changes in the membrane potential enabling the transmission of e.g. neural signals. The precise structural changes occurring in VGSCs are not completely understood, however. Now, Ayumi Sumino and Takashi Sumikama from Kanazawa University in collaboration with Katsumasa Irie from Wakayama Medical University and colleagues have succeeded in observing the structural dynamics of VGSC by means of high-speed atomic force microscopy (HS-AFM), a method capable of imaging the nanostructure and subsecond dynamics of biomolecules.
VGSCs can be in three different states: resting, inactive and active. In the latter state, Na+ ions can pass through the channel; in the resting and inactive states, which are structurally different, ions cannot pass. The basic structure of a VGSC consists of two modules: voltage sensor domains and pore domains. These domains form a square arrangement, with the ion pore at its center. An important open question is whether the voltage sensor domains dissociate from the pore domains when the channel closes.
Sumino and colleagues performed experiments on three VGSCs. One is the sodium channel of a particular bacterium (Arcobacter butzleri), the other two are mutants thereof. These three VGSCs have different voltage dependencies, with activation voltages starting at -120 mV, -50 mV and 0 mV, so that at the experimental conditions (0 mV), the VGSCs are in different states.
In order to provide insights into the structural dynamics of these three VGSCs, the researchers applied HS-AFM, a powerful technique for producing image sequences of biochemical compounds. A single AFM image is generated by laterally moving a tip just above the sample’s surface; during this xy-scanning motion, the tip’s position in the direction perpendicular to the xy-plane (the z-coordinate) will follow the sample’s height profile. The variation of the z-coordinate of the tip then produces a height map — the image of the sample. The generation of such AFM images in rapid succession then produces a video recording of the sample.
The HS-AFM results revealed that for the mutant VGSC in the resting state, the voltage sensor domains are indeed dissociated from the pore domains. Furthermore, the researchers found that the dissociated voltage sensor domains of neighboring channels connect to form pairs — this is referred to as dimerization.
The observation of the dissociation of voltage sensor domains, as well as the dimerization between pore channels, are findings that will lead to a better understanding of what causes pores to close in the resting state, and how the development of action potentials is regulated. Quoting the scientists, dimerization offers “a potential explanation for the facilitation of positive cooperativity of channel activity in the rising phase of the action potential”.
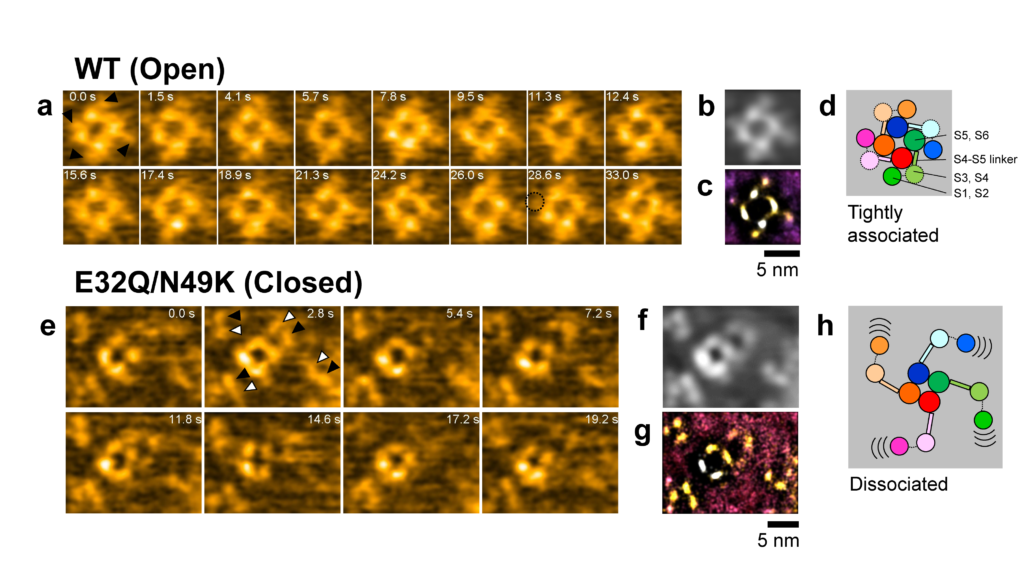
Figure 1.
HS-AFM of the Nav channels. (a and e) Snapshots of HS-AFM movies of WT (open) and E32Q/N49K (closed) channels. (b and f) Time-averaged images of (a) and (e). (c and g) Localization AFM images of (a) and (e). (d and h) Schematic illustration of results.
Glossary
Article
- Title
- Voltage sensors of a Na+ channel dissociate from the pore domain and form inter-channel dimers in the resting state
- Author
- Ayumi Sumino, Takashi Sumikama, Mikihiro Shibata, Katsumasa Irie
- Journal
- Nature Communications
- Publication date
- Dec 19, 2023
- DOI
- 10.1038/s41467-023-43347-3
- URL
- https://www.nature.com/articles/s41467-023-43347-3


HS-AFM produces a sequence of AFM images recorded in rapid succession — a video, where the time interval between frames depends on the speed with which a single image can be generated (by xy-scanning the sample). In recent years, researchers at Kanazawa University have further developed HS-AFM so that it can be applied for studying biochemical molecules and biomolecular processes in real-time. Ayumi Sumino and colleagues have now applied the method to study the structural dynamics of Na+ ion channels, revealing the dissociation of channel components and the formation of inter-channel dimers when the channel is in the resting (blocking) state.