Real-time observation of structural dynamic of influenza A hemagglutinin during viral entry
Abstract
Researchers in Kanazawa University have recently reported their study in Nano Letters regarding a high-speed atomic force microscopy study on a biological event that happens during flu virus enters infects its host cell. The real-time visualization of influenza A hemagglutinin (HA) has enhanced the understanding of fusogenic transition of HA and its interactions with host endosomes.
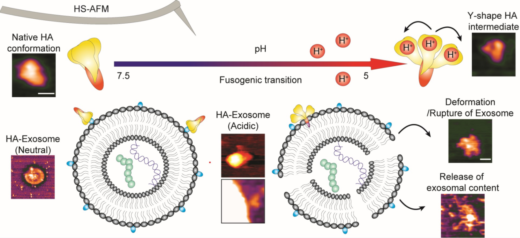
Unlike living organisms, to avoid extinction, viruses need to hijack living host machineries to generate new viruses. The devastating respiratory virus, influenza A virus, utilize its hemagglutinin (HA) proteins to search for suitable host cells. Generally, HA has two important functions: selection of host cell and viral entry. Upon attaching to host cells, Influenza A virus are brought into host cells via endocytosis. A lipid bilayer cargo, known as endosome, carries influenza A virus from cell membrane into cytoplasm of host cell. Although the environment inside endosome is acidic, influenza A virus remains alive. More strikingly, HA undergoes structural change to mediate viral membrane to fuse with host endosomal membrane to form a hole in order to release viral components. Generation of this fusion event is elaborated as fusogenic, and hence structural changes of HA needed for this event are called a fusogenic transition. The mechanism of this event has been kept in Pandora’s Box for decades despite extensive studies have been done to reveal its mystery. Now, Keesiang Lim and Richard Wong from Kanazawa University and colleagues have studied the molecular dynamic of HA using high-speed atomic force microscopy, a technique enabling real-time visualization of molecules on the nanoscale. The researchers were not only able to record the fusogenic transition of HA, but also observe its interaction with exosomes (a lipid bilayer cargo similar to endosome released by cells to outside environment).
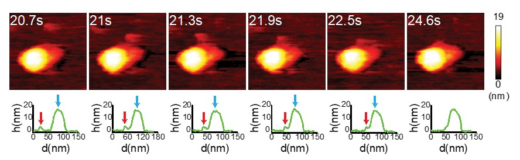
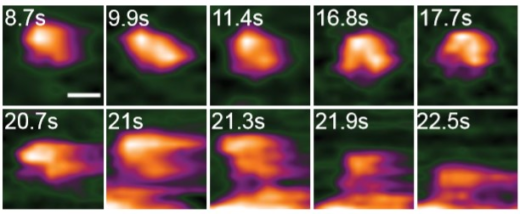
The scientists initially observed the native conformation of HA under neutral physiological buffer, a condition that resembles to a neutral condition in host cell (a pH of 7.6). In this condition, HA was appeared as an ellipsoid, which is in agreement with findings generated by other tools such as X-ray crystallography and cryo-electron microscopy. Wong and colleagues have successfully recorded the fusogenic transition, which happening when HA was exposed to an acidic environment. Their HS-AFM results illustrated a transition of HA from an ellipsoid to a Y-shape together with declination of height and circularity/roundness of HA over time. The researchers reassure the conformational change happens because a particular subunit of HA became easily to be digested by trypsin after the transition.
To study how HA can facilitate the fusion between viral membrane and host endosome membrane, Wong and colleagues let HA interacted with exosomes, a lipid bilayer cargo that mimics endosome. The HA-exosome interaction is expected to be similar to HA-endosome interaction during membrane fusion. During the interaction, conformational change of HA was found again before its docked on an exosome. Fusogenic transition releases a particular peptide, known as fusion peptide, which later inserts into the exosomal membrane, enabling the HA molecule to embed on the membrane. The scientists also found evidences that the HA-exosome interaction caused deformation or rupture of exosome, leading to a ‘leakage’ of exosomal materials.
The findings of Wong and coworkers provide important insights for the mechanism of HA-mediated membrane fusion. In addition, their work also demonstrates the advantages of HS-AFM for studying biological processes. Lim and Wong exhilaratingly commented: “This study strongly suggests that HS-AFM is a feasible tool, not only for investigating the molecular dynamic of viral fusion proteins, but also for visualizing the interaction between viral fusion proteins and their target membranes.”
Figure.
High-speed atomic force microscopy enables visualizing the fusogenic transition of HA in acidic environment, attachment to exosome membrane, and subsequent exosome rupture. Credit: Nano Letters
Background
Influenza A hemagglutinin
Influenza A hemagglutinin (HA) is a protein residing on the surface of influenza A virus (the culprit that causes ‘the flu’ or influenza), playing a key role in viral infectivity. HA’s functions include attaching influenza A virus to target cells and viral entry. After the virus attaches to its host cell, it is trapped in a lipid bilayer cargo known as endosome, and subsequently enters into the host cytoplasm. This process is called as endocytosis. Acidic environment in endosome triggers structure changes of HA to allow HA to orchestrate fusion between the viral membrane and host endosomal membrane. Finally, viral components can be released into host cells and new viruses will be made. The main target cells in human beings are typically located in the upper respiratory tract. Richard Wong from Kanazawa University and colleagues have now applied high-speed atomic force microscopy to study the fusogenic transition of HA, and the interaction of HA with lipid-bilayer membranes.
Atomic force microscopy
Atomic force microscopy (AFM) is an imaging technique in which the image is formed by scanning a surface with a very small and sharp tip. Horizontal scanning motion of the tip is controlled via piezoelectric elements, while vertical motion is converted into a height profile, resulting in a height distribution of the sample’s surface. As the technique does not involve lenses, its resolution is not restricted by the so-called diffraction limit as in X-ray diffraction, for example. In a high-speed setup (HS-AFM), the method can be used to produce movies of a sample’s structural changes in real-time, as one biomolecule can be scanned in 100 ms or less. Wong and colleagues successfully applied the HS-AFM technique to study the fusogenic transition of HA, and how it fuses with the membranes of biological particles.
Article
- Title
- High-speed AFM reveals molecular dynamics of human influenza A hemagglutinin and its interaction with exosomes
- Author
- Keesiang Lim, Noriyuki Kodera, Hanbo Wang, Mahmoud Shaaban Mohamed, Masaharu Hazawa, Akiko Kobayashi, Takeshi Yoshida, Rikinari Hanayama, Seiji Yano, Toshio Ando, and Richard W. Wong.
- Journal
- Nano Letters
- Publication date
- Aug 6, 2020
- DOI
- 10.1021/acs.nanolett.0c01755
- URL
- https://pubs.acs.org/doi/10.1021/acs.nanolett.0c01755