Control of three-dimensional multicellular dynamics by stem cell organoids and dynamics simulation

Satoru Okuda, Associate Professor, Nano Life Science Institute (WPI-NanoLSI), Kanazawa University
Integrating mechanical engineering and developmental biology
“I studied mechanical engineering for my bachelor’s degree,” says Satoru Okuda. “I learnt about the importance of mathematical simulations—mainly based on Newtonian mechanics—to determine forces acting on massive structures in order to construct safe and stable buildings. This led to me think about the factors governing the growth of tissues and organs that comprise the massive and extremely complicated structure of humans. So I decided to apply my knowledge of mechanical engineering to the world of life sciences and eventually completed a doctorate in biomechanics, simulating forces governing the dynamics of the morphogenesis of tissues, namely, how single cells form and control the development of organisms.”
The main goals of Okuda’s research at WPI-NanoLSI are prediction, designing, and manipulation of multi-cellular dynamics and understanding self-organizing mechanisms in cellular systems. The approach focusses on quantitative simulations, as widely to predict weather and design automobiles. “One of the notable unanswered questions in developmental biology is the nature and effects of mechanical forces on cells on tissue formation,” says Okuda. “This was the mystery that I decided to solve.”
The issues related to resolving this ‘mystery’ are:
- Structural hierarchy in morphogenesis—a multi-scale and multi-body problem to determine the effect of mechanical forces acting on cells.
- Robustness of morphogenesis—how do individual cells regulate the entire structure of tissues comprising organs? This suggests the existence of driving forces from cells to tissues and feedback to regulate the process.
Okuda and his colleagues have developed 3D multicellular models—cyto-element and 3D vertex models— to explore the process of cells forming tissues. “The 3D vertex model enables the prediction of 3D multi-cellular dynamics at single cell resolution,” explains Okuda. “Using dynamics models to simulate collective movement of cells in epithelial sheets we found that they are driven by force control of adherence junction.”
Other recent reports by Okuda and colleagues include in-silico tension/compression tests of epithelial luminal tissue showed the degree of freedom of cell coordination reduces tissue rigidity; application to the process of cancer invasion for theory of cell extrusion and simulations of tumorization.
Major breakthrough: Cells sense 3D tissue deformation
In 2018 Okuda and colleagues reported on their discovery that cells sense 3D tissue deformation using a pluripotent stem cell–derived optic-cup organoid model. “Our new mathematical framework predicted for simulating general 3D multi-cellular morphogenesis at single cell resolution,” says Okuda. “In our approach we combined mechanics-based simulations and brain organoid and found that individual cells sense mechanical strain to modulate 3D tissue deformation. This is a breakthrough in developmental biology because this shows that mechanical forces do indeed act as ‘feedback regulators’ to ensure robust organogenesis.”
Okuda and his research team will continue to combine mathematical modeling, organoid culture, active perturbation systems, and quantitative measurements to demystify the fundamental mechanisms governing self-organizing in multi-cellular systems.
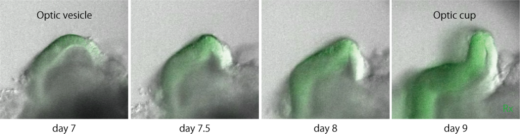
Pluripotent cell-derived optic cup formation

Optic-cup recapitulated by computational simulation
Computational simulation of optic-cupformation
References
- Satoru Okuda, Erina Kuranaga, Katsuhiko Sato, “Apical junctional fluctuations lead to cell flow while maintaining epithelial integrity,” Biophysical Journal 116 (6): 1159-1170 (2019)
- Satoru Okuda, Nozomu Takata, Yuiko Hasegawa, Masako Kawada, Yasuhiro Inoue, Taiji Adachi, Yoshiki Sasai, Mototsugu Eiraku, “Strain-triggered mechanical feedback in self-organizing optic cup morphogenesis,” Science Advances 4(11): eaau1354 (2018)
- Satoru Okuda, Takashi Miura, Yasuhiro Inoue, Taiji, Adachi, Mototsugu Eiraku, “Combining Turing and 3D vertex models reproduces autonomous multicellular morphogenesis with undulation, tubulation, and branching,” Scientific Reports 8: 2386 (2018)
- Atsushi Hashimoto, Atsuki Nagao, Satoru Okuda, “Topological graph description of multicellular dynamics based on vertex model,” Journal of Theoretical Biology 437: 187-201 (2018)

