Drug metabolism and toxicology: Monitoring interactions of enzymes for drug development and next generation pharmacotherapy
Role of drug-metabolizing enzymes that determine differences in drug response and toxicity of individuals

Miki Nakajima, Professor, Nano Life Sciences Institute (WPI-NanoLSI), Kanazawa University
Medically prescribed drugs are ubiquitous, and it is difficult to imagine a world without them. Such pharmaceutical drugs (drugs) are chemicals used to treat or prevent ailments ranging from diabetes to cancer and headaches to cardiac malfunction.
The body reacts to drugs in the same way as with all foreign objects, by trying to eliminate them to protect the body. Typically, tablets or capsules of a drug are dissolved in the stomach and their components are absorbed in the intestine then delivered to the liver, which detoxifies the compounds that then find their way into the blood circulation system and target tissues, and finally the remnants are excreted.
“The series of four steps when a drug is taken are absorption, distribution, metabolism and excretion, that is ADME,” says Miki Nakajima. “This area of research—known as pharmacokinetics or drug metabolism—is one of the main areas of my studies. I want to clarify the mechanisms governing inter or intra-individual variability in drug response and toxicity or drug-drug interactions for the development of drugs and optimized pharmacotherapy. So it is important to know how the human body responds to drugs as they diffuse through it.” Figure 1 is an illustration of the process of ADME as a drug diffuses through the body after administration.

Fig. 1. Variation of drug diffusion in the body over with time
Importantly, the liver degrades drugs via two main so-called metabolic processes: Phase I metabolism refers to the chemical transformation of drugs to hydrophilic compounds that are readily eliminated by the kidneys; Phase II metabolism is a step to increase the hydrophilicity of chemicals further or to detoxify the formed reactive metabolites.
“In our research we are focusing cytochrome P450 enzymes that govern Phase I metabolism in humans,” explains Nakajima. “These drug-metabolizing enzymes determine the observed differences in drug response and toxicity between individuals taking the same drugs.”
Recently, Nakajima and colleagues discovered that human CYP2A6—a cytochrome P450—is regulated by microRNA. “We found that CYP2A6 is post-transcriptionally regulated by miR-126* and that CYP2A7 affects the expression of CYP2A6 by competing for miR-126* binding. These findings are important for the development of anti-cancer drugs and carcinogens related to tobacco.” [1].
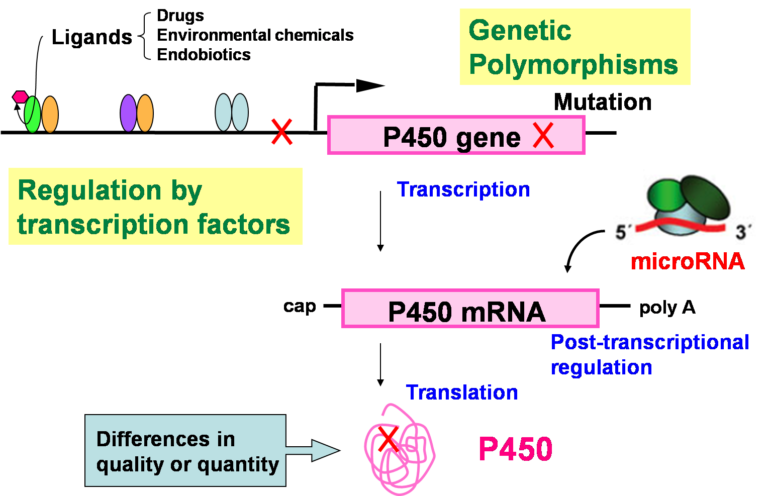
Fig. 2. Factors causing interindividual differences in the expression and/or function of human P450 enzymes.
Plans for research at the WPI-NanoLSI include using AFM technology to monitor interactions of P450 and P450 reductase on lipid membrane and related processes; development of supramolecules capable of capturing reactive metabolites; and analysis of the intervention of exosomes in drug-induced toxicity caused by reactive metabolites.
Research highlights
In their 2018 paper published in Pharmacology & Therapeutics [2] Nakajima and colleagues describe their research on anti-cancer drug target molecules and their findings that A-to-I RNA editing influences drug responsiveness.
As internationally acclaimed publication, Nakajima and her colleagues in Japan, reported the first observation that microRNAs regulate both essential genes for physiologic events and drug-metabolizing enzymes [3].
References
- M. Nakano et al, “CYP2A7 pseudogene transcript affects CYP2A6 expression in human liver by acting as a decoy for miR-126*”, Drug Metab Dispos, 43, 703-712 (2015)
DOI: 10.1124/dmd.115.063255 - M. Nakano and M. Nakajima, “Significance of A-to-I RNA editing of transcripts modulating pharmacokinetics and pharmacodynamics”, Pharmacology & Therapeutics 181, 13-21 (2018).
DOI: 10.1016/j.pharmthera.2017.07.003 - Y. Tsuchiya et al, “MicroRNA regulates the expression of human cytochrome P450 1B1”, Cancer Res 66, 9090-9098, (2006).
DOI:10.1158/0008-5472.CAN-06-1403

