From Hydration Layers to Nanoarchitectures: Water’s Pivotal Role in Peptide Organization on 2D Nanomaterials
Researchers at the Nano Life Science Institute (WPI-NanoLSI), Kanazawa University, report in Small, a weekly peer-reviewed scientific journal covering nanotechnology, published by Wiley-WCH, Germany, how short peptides self-assemble linearly on atomically-thick solid surfaces, such as graphite and MoS₂. The research addresses a longstanding challenge in materials science: understanding the complex, sequence-specific interactions between peptides and solid substrates, and the critical role of local hydration structures in guiding nanoarchitecture formation. This work offers new strategies for integrating biomolecules with advanced materials in future bioelectronics and sensor devices.
Biotechnological applications typically exploit the properties and functionalities of biological molecules. For practical biotechnology devices, it is essential to assemble biomolecules on non-biological substrates. Fortunately, specifically designed biomolecules can achieve structural ordering and enable potent bionano-hybrid applications through spontaneous self-organization on such substrates. Still, the mechanisms behind on-substrate self-assembly are not well understood. Indeed, biomolecular self-assembly is not only governed by the structural features of the molecules and the substrate, but also by the solvent in which the process takes place. Recently, a team of researchers led by Ayhan Yurtsever, Takeshi Fukuma, and Linhao Sun from Kanazawa University, in collaboration with Yuhei Hayamizu at the Institute of Science Tokyo and Mehmet Sarikaya from DMXi Dentomimetix, Inc., Washington, USA, performed a detailed investigation of the assembly process of peptides on inorganic substrates; by employing state-of-the-art visualization techniques and computer simulations led by Fabio Priante, and Adam S. Foster from Aalto University, Finland, the team provided new insights into the mechanisms involved, particularly highlighting the critical role of water as the solvent (Figure 1).
The researchers designed short and simple dipeptides, mainly consisting of alternating amino acids with aromatic side groups, tyrosine (Y) and histidine (H) (Figure 2). The former is hydrophobic residue, which provides water-repelling environment, whereas the latter is hydrophilic, creating a water-attractive background. By varying the number of repeating YH units (3, 4, and 5), the team systematically investigated how these peptides form linear, crystalline structures aligned with the underlying atomic lattice on 2D crystallographic interfaces such as graphite and MoS₂.
Using frequency modulated atomic force microscopy (FM-AFM), a technique for visualizing surfaces, the team visualized the assembly of Tyr-His (YH) dipeptides with tandem repeats on cleaved graphite and MoS₂. Their findings showed that peptides adopt fully extended, linear conformations aligned with the specific crystallographic orientations of the underlying substrate. Remarkably, the measured lengths of these assemblies, including their hydration layers, matched the peptides’ unfolded states (Figure 3). These findings highlight the critical interplay between aromatic interactions and solvation effects in guiding peptide self-assembly. Yurtsever and colleagues emphasize that water molecules not only facilitate intermolecular hydrogen bonding but also provide the conformational flexibility necessary for peptides to adapt during assembly, thereby enabling subtle structural adjustments that support the overall assembly process. Computer simulations further revealed that hydrophobic interactions between the peptide molecules and the substrate, along with specific intermolecular interactions, stabilize the observed dipeptide arrangements.
The scientists then performed advanced 3D-AFM measurements to probe the 3D structure of the water around the peptide assemblies. They discovered that peptide-water interactions lead to the formation of heterogeneous hydration shells, which encapsulate the peptide assemblies and create specific binding pockets. These features are crucial for selective molecular recognition and could mediate interactions with other biomolecules. Molecular dynamics simulations corroborated these findings, offering detailed microscopic insights into the hydrogen-bond network that shapes the structure and stability of the hydration layer.
The study by Yurtsever and colleagues provides insights that open new avenues for the rational design and functional control of peptide-based hybrid materials, offering a robust platform for biofunctionalization in biomedical and bionanotechnology applications, including biosensors and bioelectronics. The well-ordered peptide lattices could serve as templates for organizing inorganic nanoparticles with sub-nanometer precision, enabling the exploration of quantum mechanical effects. Moreover, the spatial arrangement of peptide side chains may facilitate the creation of catalytically active sites that mimic natural enzymes, as well as support the immobilization of biomolecules for molecular recognition studies and high-performance catalytic interfaces in electrochemical applications.
The teams are currently focused on further unraveling the local hydration structures surrounding solid-binding peptides, providing deeper insight into how hydrophobic and hydrophilic sequences influence water organization at interfaces and advancing the understanding of the molecular mechanisms underlying peptide assembly on solid surfaces.
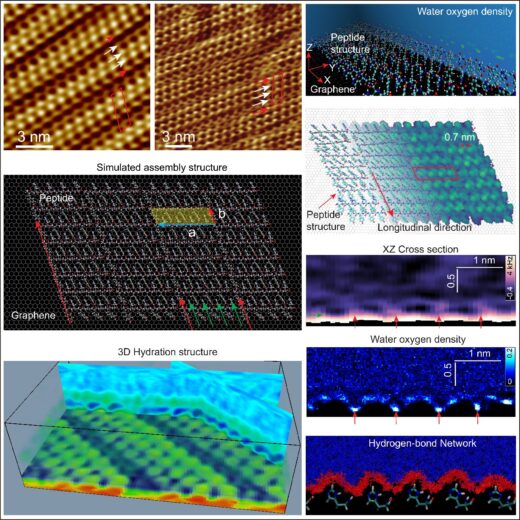
Figure 1: Molecular assembly structure of YH dipeptides on graphite and MoS₂; associated three-dimensional hydration structures surrounding peptide assemblies; simulated assembly structure and water density distribution near the assembled peptides.
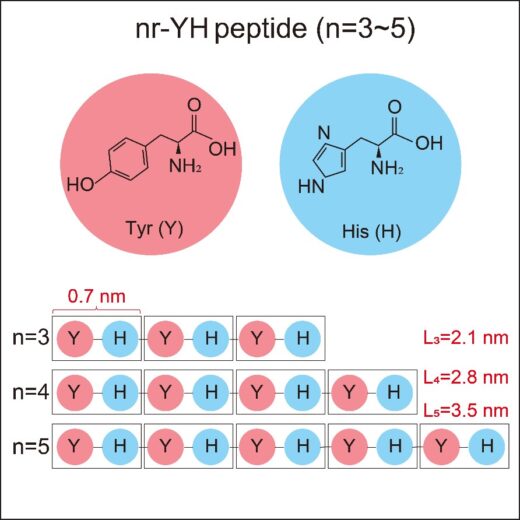
Figure 2: Molecular characteristics of nr-YH peptides (n=3, 4, 5). The molecular structure of the peptide contains two amino acids: Tyrosine, with a phenol side chain, and Histidine with imidazole ring. The schematic represents the sequences of the peptides and their lengths in their unfolded conformations.
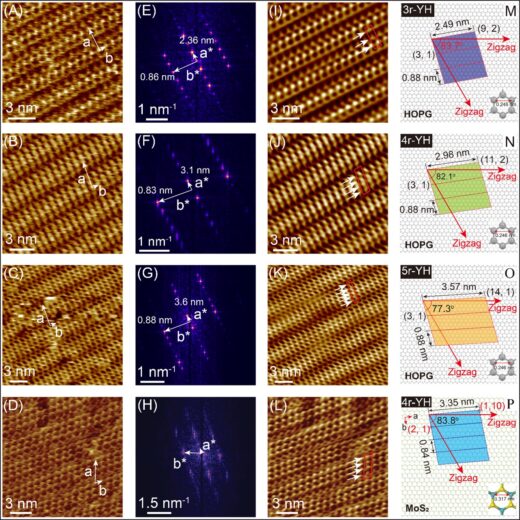
Figure 3. Structural characteristics of the self-assembled nr-YH peptide on graphite and molybdenite. (A to C) FM-AFM-recorded molecular resolution images of the self-assembled 3r-, 4r-, and 5r-YH peptides forming 2D lattices on graphite in water and that of 4r-YH on MoS₂ (D). (E to H) The corresponding FFT pattern obtained from the peptide assemblies in panels (A-D). The reconstructed images in (I-L) are obtained by executing inverse 2D FFT spectra (E-H). The white arrows show bright contrast features in a given peptide lattice, the number of which is consistent with the repeating YH units for each peptide assembly. (M-P) Crystallographic orientation relationships between the peptide crystals and the solid lattice obtained through matching the molecular lattice of the peptide and the atomic lattice of the solids that give chiral relationship for each peptide. All images were recorded by in situ FM-AFM using 1.5 µM peptide solutions.
All credits, including the eye-catching image, © 2025 Yurtsever, et al., Small published by Wiley-VCH GmbH
Article
- Title
- Supramolecular Assembly and Interfacial Hydration of Tandem Repeat Dipeptides on 2D Nanomaterials: Insights from 3D-AFM Measurements and MD Simulations
- Author
- Ayhan Yurtsever, Fabio Priante, Linhao Sun, Marie Sugiyama, Kazuki Miyata, Keisuke Miyazawa, Sanhanut Kesornsit, Katsuhiro Maeda, Adam S. Foster, Yuhei Hayamizu, Mehmet Sarikaya, and Takeshi Fukuma
- Journal
- Small
- Publication date
- Jun 11, 2025
- DOI
- 10.1002/smll.202501785
- URL
- https://onlinelibrary.wiley.com/doi/10.1002/smll.202501785

