Small but mighty: Identifying nanosized molecules using atomic force microscopy
In a study recently published in the journal Applied Materials and Interfaces researchers from Kanazawa University use advanced atomic force microscopy to accurately recognize tiny cellular biomolecules
Biologists rely on a wide range of microscopy techniques to visualize biomolecules within biological cells. High-speed atomic force microscopy (HS-AFM) is one such example in which a sharp tip attached to a sensor is used for visualizing cells. Specifically, as the AFM tip scans the surface of a molecule, the pattern of signals it generates enables researchers to visualize the molecule’s topography. However, recognizing individual biomolecules using HS-AFM is still in its infancy. Now, researchers at Kanazawa University report on an innovative method to facilitate this by tweaking the structures of AFM tips.
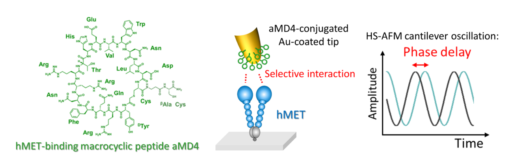
The research team, led by Mikihiro Shibata and Leonardo Puppulin at the WPI Nano Life Science Institute Kanazawa University (NanoLSI), characterized a protein found on human cells known as the hepatocyte growth factor receptor (hMET). The researchers first attached aMD4, a synthetic molecule that latches onto hMET, to the HS-AFM tip using different sized linkers. Patterns of connections between this modified tip and single molecules of hMET were subsequently investigated.
When hMET on a mica surface (a material typically used in HS-AFM studies) was exposed to the tip, interactions between aMD4 and the external surface of hMET were indeed observed. However, when multiple molecules of aMD4 and hMET were used, it was found that shorter and more flexible linkers enabled aMD4 to interact with two adjacent hMET molecules bringing them closer together. This observation posits practical applications in the laboratory—biologists can potentially mimic the binding of two cell surface proteins together which often leads to the induction of cellular processes.
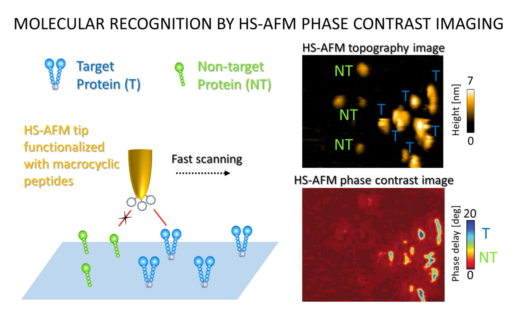
Next, the specificity of this tip in molecule recognition was examined. hMET and its mouse form are very similar in structure. However, the mouse form does not bind to aMD4. Thus, when the aMD4-loaded tips were exposed to both forms of the protein, activity was observed only with the human form. This technique could therefore be useful in the selection of specific biomolecules from a heterogenous mix as is typically seen on the cell surface. Lastly, the modified HS-AFM technique was applied when hMET was bound to a lipid surface mimicking the structural composition of cell membranes (its natural home). Similar interactions were observed in this milieu suggesting the potential for protein recognition on living cells.
“The versatility of macrocyclic peptides in detecting unlimited types of membrane receptors with high selectivity and the fast imaging by HS-AFM broaden the range of future applications of this method for molecular recognition without labeling,” concludes the team. The applications of this method in cell biology are vast, including tracking the movement and concentration of cell-surface proteins and their subsequent cellular effects.
Image
Figure 1. A graphical representation of the molecular structure of aMD4 (macrocyclic peptide) and the interaction with hMET when attached to the HS-AFM tip. The selective interaction induces peculiar delay of the cantilever oscillation.
Figure 2. HS-AFM images showing increased activity when the ligand-bound tip interacted with the human MET protein (target, T) and not the mouse version of the protein (non-target, NT).
Background
High-speed atomic force microscopy: Atomic force microscopy is a tool used in structural biology to investigate the topography (or 3D shape) of a molecule. High-speed atomic force microscopy (HS-AFM) is an advanced form wherein these topographical images are taken very quickly, enabling a dynamic visualization. Thus, the structure of molecules in real-time can be captured. However, if scientists want to identify and capture specific biomolecules using HS-AFM, they must tag these molecules beforehand. This is often a cumbersome and nonspecific process. The technique shown in this study, however, makes it easy to identify proteins by manipulating only the HS-AFM tip instead.
Directory of Researcher
Article
- Title
- Macrocyclic Peptide-Conjugated Tip for Fast and Selective Molecular Recognition Imaging by High-Speed Atomic Force Microscopy
- Author
- Leonardo Puppulin, Daiki Kanayama, Naohiro Terasaka, Katsuya Sakai, Noriyuki Kodera, Kenichi Umeda, Ayumi Sumino, Arin Marchesi, Wei Weilin, Hideo Tanaka, Takeshi Fukuma, Hiroaki Suga, Kunio Matsumoto, Mikihiro Shibata
- Journal
- ACS Appl. Mater.
- Publication date
- Nov 12, 2021
- DOI
- 10.1021/acsami.1c17708
- URL
- https://pubs.acs.org/doi/10.1021/acsami.1c17708