Insight into structural remodeling of the FlhA ring responsible for bacterial flagellar type III protein export
Abstract:
The bacterial flagellum is a supramolecular machine composed of the basal body, hook and filament. Its building block proteins are synthesized and transported to respective worksites in a given order to construct the flagellum. FlhA, a component of the flagellar type III protein export apparatus, coordinates export and assembly of the building block proteins. We observed the C-terminal domain of FlhA (FlhAC) and its mutants using high-speed AFM, and discovered that a flexible linker of FlhAC is essential both in ring formation of FlhAC and in switching an exporting protein from the hook protein to the filament protein upon completion of the hook assembly.
Background
The bacterial flagellum consists of the basal body, the hook and the filament, assembled in this order from the bacterial cell surface (Figure 1). A flagellum is constructed such that the building block proteins are assembled with extreme precision at the most distal part, the farthest from the cell, of an assembling flagellum. A specific flagellar protein export apparatus*1 at the base of a flagellum is responsible for exporting just a sufficient number of necessary component (building block) proteins, to the assembly site, depending on the assembly process (Figure 2). The flagellar protein export apparatus exports from time to time FliK, a kind of molecular ruler protein, in order to monitor the assembly taking place. Upon completion of the hook structure to about 55 nm in length, FliK provides information on the hook length for a component protein FlhB of the flagellar protein export apparatus. This changes the substrate specificity of the flagellar protein export apparatus to stop hook assembly and to initiate filament formation. However, the mechanism for switching the proteins to be exported by the flagellar protein export apparatus was unknown.
Results
This study was performed by a team consisting of researchers from several Japanese institutions including Kanazawa University. The team focused on FlhA, which forms a nonameric ring structure in the flagellar protein export apparatus, and carried out dynamic structure analyses of FlhAC (carboxyl-terminal cytoplasmic domain of FlhA) using a high-speed atomic force microscope*2. They demonstrated that FlhAC forms a nonameric ring; a flexible linker, which is an extension of the transmembrane domain of FlhA, is essential for the cooperative nonameric ring formation. Point mutation of an amino acid residue of FlhAC important in the nonameric ring formation prevents not only ring formation but also switching of the substrate specificity of the flagellar protein export apparatus. These results suggest that, upon completion of the hook structure, the flexible linker of FlhA binds to adjacent FlhAC in a cooperative manner, significantly changing the overall structure of the FlhAC nonameric ring. It is also suggested that, as a consequence, export of the hook protein is stopped and that of the filament protein is initiated (Figure 3).
The present study has successfully visualized the switching mechanism of the substrate specificity of the flagellar protein export apparatus, which remained a mystery for a long time, and has shed light on the switching mechanism itself. It is expected that the mechanism of hook completion signaling*3 complex formation, which exists only transiently upon completion of the hook structure, would become clear together with the functional mechanisms of this signaling complex.
Significance and future prospects
Antibacterial agents are often used for preventing bacterial infection. However, because of the emergence of multidrug-resistant bacteria, an increasing number of infectious diseases are becoming difficult to treat. Thus it is an urgent task to develop novel means of prevention of infectious diseases. The type III secretion apparatus*4 that pathogenic bacteria use for infection is specific to bacteria but is not necessary for bacterial survival. If a drug were to be discovered that inactivates the type III secretion apparatus only, it could abolish only the pathogenicity of infectious bacteria. Since the type III secretion apparatus is similar to the flagellar protein export apparatus in terms of both its function and structure, the outcome of this study is expected to contribute to the development of new technology to regulate bacterial infectious diseases, by enabling new drug screening and discovery targeting the FlhA homologue, a structural component of the type III secretion apparatus.
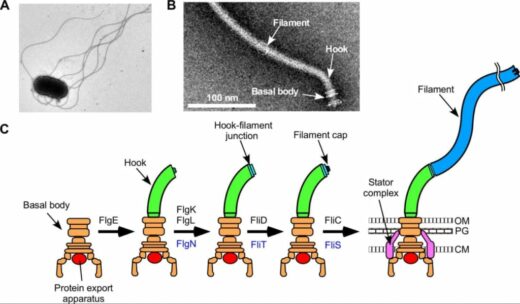
Figure 1. Structure of a bacterial flagellum.
(A) Electron microscopic image of a Salmonella cell.
(B) Electron microscopic image of a flagellum isolated and purified from Salmonella.
(C) Process of formation of a flagellum. OM, outer membrane; PG, peptideglycan layer; M, cytoplasmic membrane.
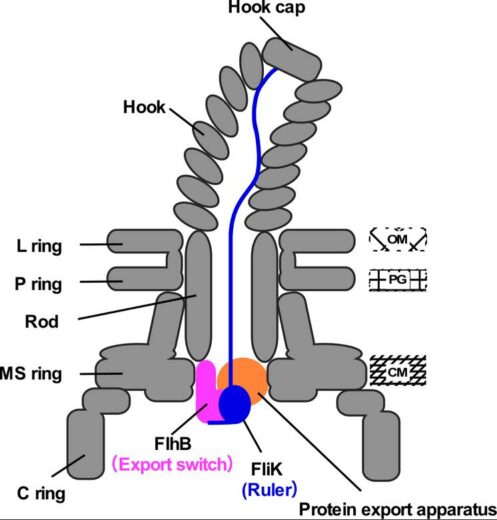
Figure 2. Schematic illustration of the transiently-formed hook completion signaling complex.
FliK, exported from the cell from time to time, functions as a molecular ruler to measure the length of the hook. When the hook attains a length of 55 nm, the C-terminal domain of FliK interacts with FlhB, a switch for protein export, and as a consequence, the substrate specificity of the protein export apparatus changes.

Figure 3. Ring formation of FlhA.
(A) Crystal structure of FlhA. It shows the linker binding to adjacent FlhAC molecule in crystal.
(B) Typical high-speed AFM images of FlhAC ring. Left, wildtype FlhAC. Right, linker-deficient mutant. Linker-deficiency inhibits the ring formation to a considerable extent.
(C) Hypothetical model for cooperative remodeling of the FlhAC ring structure. When the linker of FlhA binds to an adjacent FlhAC subunit, the FlhAC ring structure is changed. As a consequence, export of the hook protein is stopped and export of the filament protein is initiated.
Glossary
Article
- Title
- Insight into structural remodeling of the FlhA ring responsible for bacterial flagellar type III protein export
- Author
- Naoya TERAHARA, Yumi INOUE, Noriyuki KODERA, Yusuke V. MORIMOTO, Takayuki UCHIHASHI, Katsumi IMADA, Toshio ANDO, Keiichi NAMBA and Tohru MINAMINO
- Journal
- Science Advances
- Publication date
- Apr 26, 2018
- DOI
- 10.1126/sciadv.aao7054
- URL
- http://advances.sciencemag.org/content/4/4/eaao7054
Funder
JSPS KAKENHI grants JP15H04360, JP15K14498, JP15H05593, JP15H03540, JP15H02386, JP24227005, JP25000013, and JP26293097; Technology KAKENHI JP26115720, JP15H01335, JP26119003, JP24117004 and JP15H01640; JST grants JPMJPR13L4 and JPMJCR13M1.

