Genetic engineering mechanism visualized
Abstract:
 Researchers at Kanazawa University and the University of Tokyo report in Nature Communications the visualization of the dynamics of ‘molecular scissors’ — the main mechanism of the CRISPR-Cas9 genetic-engineering technique.
Researchers at Kanazawa University and the University of Tokyo report in Nature Communications the visualization of the dynamics of ‘molecular scissors’ — the main mechanism of the CRISPR-Cas9 genetic-engineering technique.
One of the techniques used in genetic engineering — the process of artificially modifying the genome of a living organism — involves the so-called CRISPR-Cas9 nuclease system. Using this system, a cell’s DNA can be cut at a desired site, where genes can be deleted or added. Selection of the site to be cut is done by a ‘guide RNA’ molecule bound to the Cas9 protein. Now, a team of researchers led by Mikihiro Shibata from Kanazawa University and Osamu Nureki from the University of Tokyo has visualized the dynamics of the CRISPR-Cas9 complex, in particular how it cuts DNA, providing valuable insights into the CRISPR-Cas9-mediated DNA cleavage mechanism.
For their visualization studies, the scientists used high-speed atomic-force microscopy (HS-AFM), a method for imaging surfaces. A surface is probed by moving a tiny cantilever over it; the force experienced by the probe can be converted into a height measure. A scan of the whole surface then results in a height map of the sample. The high-speed experimental set-up of Shibata and colleagues enabled extremely fast, repeated scans — convertible into movies — of the biomolecules taking part in the molecular scissoring action.
First, the scientists compared Cas9 without and with RNA attached (Cas9–RNA). They found that the former was able to flexibly adopt various conformations, while the latter has a fixed, two-lobe structure, highlighting the conformational-stabilization ability of the guide RNA. Then, Shibata and colleagues looked at how the stabilized Cas9–RNA complex targets DNA. They confirmed that it binds to a pre-selected protospacer adjacent motif (PAM) site in the DNA. A PAM is a short nucleotide sequence located next to the DNA’s target site, which is complementary to the guide RNA.
The research team’s high-speed movies further revealed that targeting (‘DNA interrogation’) is achieved through 3D diffusion of the Cas9–RNA complex. Finally, the researchers managed to visualize the dynamics of the cleavage process itself: they observed how the region of ‘molecular scissors’ undergoes conformational fluctuations after Cas9–RNA locally unwinds the double-stranded DNA (Figure 2).
The work of Shibata advances our understanding of the CRISPR-Cas9 genome-editing mechanism. In the words of the researchers: ” … this study provides unprecedented details about the functional dynamics of CRISPR-Cas9, and highlights the potential of HS-AFM to elucidate the action mechanisms of RNA-guided effector nucleases from distinct CRISPR-Cas systems.”
[Background]
CRISPR-Cas9
CRISPR, short for “clustered regularly interspaced short palindromic repeats”, refers to a set of bacterial DNA sequences containing fragments of the DNA of viruses having earlier attacked the bacteria. These fragments are used by the bacteria to prevent further attacks by the same viruses. “Cas” refers to CRISPR-associated genes; “Cas9” is a CRISPR-associated protein with two nuclease domains (A nuclease is an enzyme capable of cleaving nucleic acids, organic molecules present in DNA and RNA).
In recent years, a genetic-engineering technique where a CRISPR-Cas9 complex acts as ‘molecular scissors’ has been developed; the Cas9 nuclease binds to a guide RNA molecule that contains information about the DNA site to target. Using high-speed atomic force microscopy, Mikihiro Shibata from Kanazawa University and colleagues have now studied the dynamics of the CRISPR-Cas9 complex in great detail.
Atomic force microscopy
Atomic force microscopy (AFM) is an imaging technique in which the image is formed by scanning a surface with a very small tip. Horizontal scanning motion of the tip is controlled via piezoelectric elements, while vertical motion is converted into a height profile, resulting in a height distribution of the sample’s surface. As the technique does not involve lenses, its resolution is not restricted by the so-called diffraction limit. In a high-speed setup, AFM can be used to produce movies of a sample’s evolution in real time. High-speed AFM has been used successfully to study protein dynamics, for example myosin V walking on an actin filament, the photo-induced conformational change of bacteriorhodopsin, and the degradation of cellulose. Shibata and colleagues have now applied the high-speed AFM technique for visualizing the dynamics of DNA cleavage by CRISPR-Cas9.
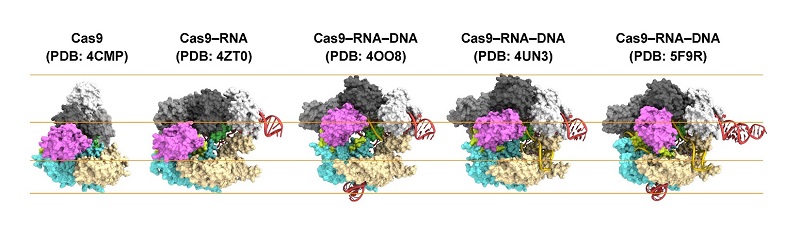
Figure 1: Structures of Cas9.
From left to right: Cas9 alone (apo-Cas9), Cas9 bound to RNA (Cas9–RNA), Cas9–RNA bound to its single-stranded DNA target (Cas9–RNA–DNA), Cas9–RNA bound to a partial DNA duplex (Cas9–RNA–DNA) and Cas9–RNA bound to its double-stranded DNA target (Cas9–RNA–DNA).
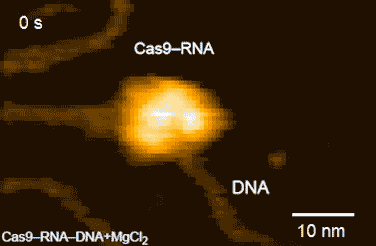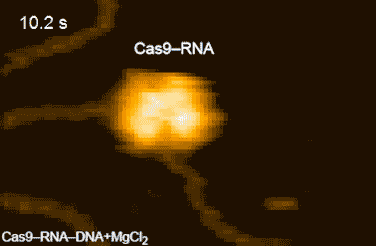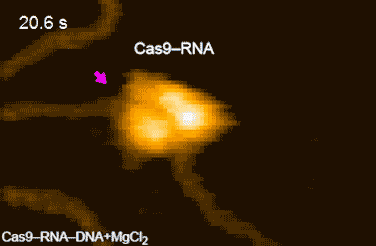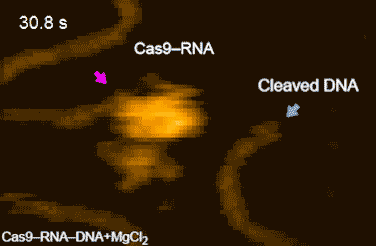
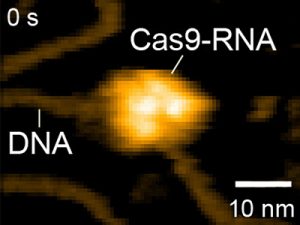
Figure 2: HS-AFM movies of DNA cleavage by Cas9–RNA.
Fluctuations of the nuclease domain are indicated by magenta arrows. The cleavage products released from Cas9–RNA are indicated by blue arrows. ↑
Article
- Title
- Real-space and real-time dynamics of CRISPR-Cas9 visualized by high-speed atomic force microscopy
- Author
- Mikihiro Shibata, Hiroshi Nishimasu, Noriyuki Kodera, Seiichi Hirano, Toshio Ando, Takayuki Uchihashi, and Osamu Nureki
- Journal
- Nature Communications
- Publication date
- Nov 10, 2017
- DOI
- 10.1038/s41467-017-01466-8
- URL
- http://doi.org/10.1038/s41467-017-01466-8
Funder
The Kao Foundation for Arts and Science, The Brain Science Foundation, JST/PRESTO, JST/CREST, The Basic Science and Platform Technology Program for Innovative Biological Medicine from AMED, JSPS KAKENHI

