BioAFMviewer software for simulated atomic force microscopy of biomolecules
Abstract
Atomic force microscopy (AFM) allows to obtain images and movies showing proteins at work, however with limited resolution. The developed BioAFMviewer software opens the opportunity to use the enormous amount of available high-resolution protein data to better understand experiments. Within an interactive interface with rich functionality, the BioAFMviewer computationally emulates tip-scanning of any biomolecular structure to generate simulated AFM graphics and movies. They greatly help in the interpretation of e.g., high-speed AFM observations.
Nowadays nanotechnology allows to observe single proteins at work. Under atomic force microscopy (AFM), e.g., their surface can be rapidly scanned, and functional motions monitored, which is of great importance for applications in all fields of Life science. The analysis and interpretation of experimental results remains however challenging because the resolution of obtained images or molecular movies is far from perfect. On the other side, high-resolution static structures of most proteins are known, and their conformational dynamics can be computed in molecular simulations. This enormous amount of available data offers a great opportunity to better understand the outcome of resolution-limited scanning experiments.
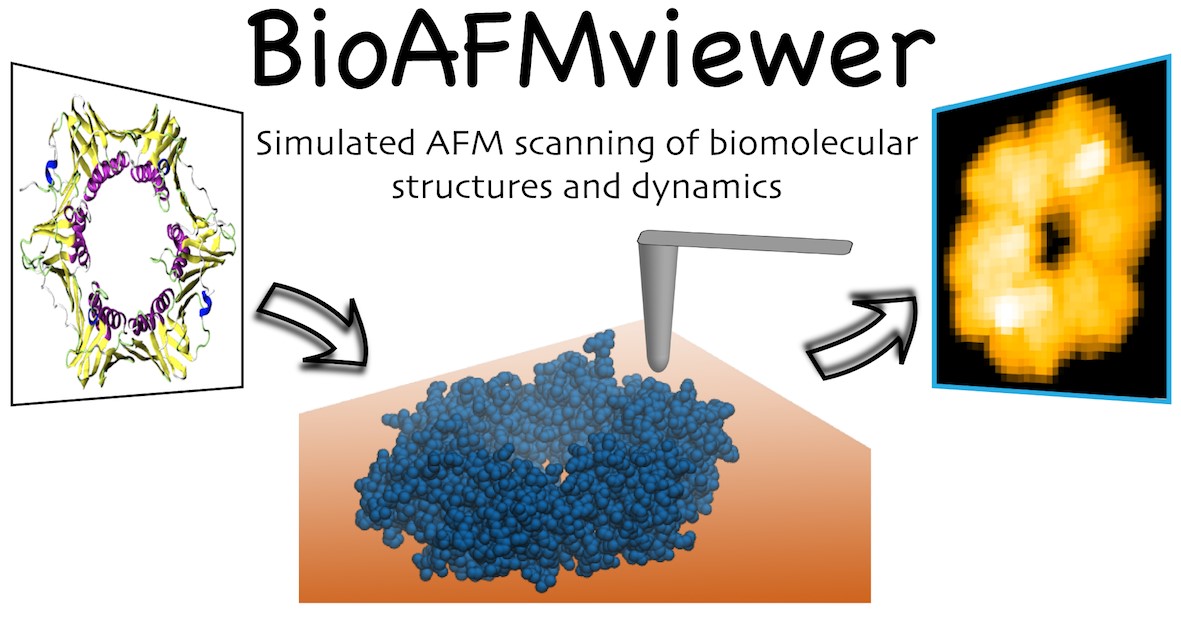
The developed software provides the computational package towards this goal. The BioAFMviewer computationally emulates AFM scanning of any biomolecular structure to generate graphical images that resemble the outcome of AFM experiments (see Fig. 1). This makes the comparison of all available structural data and computational molecular movies to AFM results possible. The BioAFMviewer has a versatile interactive interface with rich functionality. An integrated 3D viewer visualizes the molecular structure while synchronized computational scanning with adjustable tip-shape geometry and spatial resolution generates the corresponding simulated AFM graphics. Obtained results can be conveniently exported as images or movies.
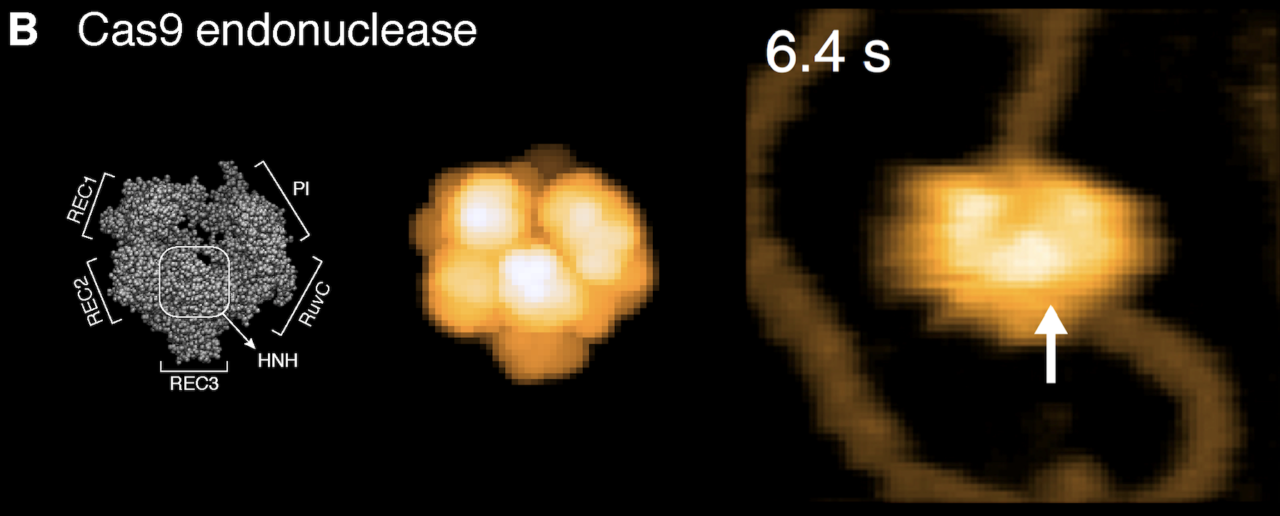
To demonstrate the great potential of simulated AFM scanning in supporting the analysis and interpretation of experimental data, the authors provide several applications to high-speed AFM observations of proteins. As an example, for the gene scissor related CRISPR-associated protein 9 endonuclease (Cas9), simulated scanning allows to disambiguate the domain arrangement seen in the high-speed AFM image and clarify their orientation with respect to the bound nucleotide strand (see Fig. 2). The authors furthermore demonstrate how the BioAFMviewer can transform molecular movies of proteins, obtained for example from molecular modeling, into corresponding simulated AFM movies. Therefore, simulated AFM experiments are possible and can be compared to the movies recorded in high-speed AFM experiments to better understand the resolution-limited observed conformational dynamics.
The BioAFMviewer has a user-friendly interface and no expert knowledge is required to use it. The software is already used by AFM groups worldwide, and it is expected to become a standard platform used by the broad community of Bio-AFM experimentalists. Beyond that, it also provides the interface for researchers from the fields of computational biology and bioinformatics to foster their interdisciplinary collaborations.
The BioAFMviewer software package is currently available for the Windows 10 operating system. A free download is provided on the project website www.bioafmviewer.com, where also future updates will become available.
The BioAFMviewer software project was initiated by Holger Flechsig, who is an Assistant Professor of the Nano Life Science Institute at Kanazawa University, Japan, where world-leading AFM experiments of biological matter are performed. The first author, Romain Amyot has developed the software package and continues to work on future applications. Besides that, he also performs high-speed AFM experiments of proteins as a postdoctoral researcher at the Aix-Marseille University, France.
Figure1 BioAFMviewer
Schematic illustration of the BioAFMviewer functionality. Computational AFM scanning transforms any biomolecular structure provided in the standard PDB file format into a corresponding simulated AFM graphics.
Figure2 Simulated scanning.
Comparison of simulated AFM graphics to high-speed AFM experiments for the Cas9 endonuclease protein. The simulated AFM graphics (middle) of the molecular structure (left) very well agrees with the experimental image obtained from high-speed AFM (right). The experimental image is adopted from the publication: M. Shibata et al. Nat. Commun. 8: 1430 (2017).
Article
- Title
- BioAFMviewer: An interactive interface for simulated AFM scanning of biomolecular structures and dynamics
- Author
- Romain Amyot, Holger Flechsig
- Journal
- PLoS Computational Biology
- Publication date
- Nov 18, 2020
- DOI
- 10.1371/journal.pcbi.1008444
- URL
- https://doi.org/10.1371/journal.pcbi.1008444
Funder
This work was supported by World Premier International Research Center Initiative (WPI), MEXT, Japan.